Haoran Xue, Ph.D. Candidate
Department of Ecology & Evolutionary Biology
University of Toronto
25 Willcocks Street
Toronto, Ontario, Canada
M5S 3B2
Office: Earth Science Building 2043
E-mail: haoran.xue@mail.utoronto.ca
Degrees
B.Sc. Biological Sciences (2017)
Peking University, China

Current Position
Ph. D. candidate, Department of Ecology & Evolutionary Biology, University of Toronto
Project: The molecular genetic architecture of tristyly
Co-supervised by Spencer C.H. Barrett and Stephen I. Wright
Research Interests
I have broad interests in ecology and evolutionary biology. I am particularly interested in the evolutionary genetics of plant mating systems. For my Ph.D. thesis, I am using approaches in molecular biology and bioinformatics to study the genetic architecture of the well-known floral polymorphism tristyly.
The polymorphism functions to promote cross-fertilization, specifically disassortative (intermorph) mating, and to reduce sexual interference and gametic wastage in plant populations and has evolved independently in six flowering plant families. I am studying this polymorphism in the annual aquatic plant Eichhornia paniculata (Pontederiaceae), which is native to N.E. Brazil, the Caribbean (Cuba and Jamaica) and Central America (Mexico and Nicaragua).
-

- Eichhornia paniculata (long-styled morph)
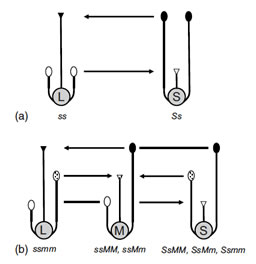
The three floral morphs that characterize the tristylous syndrome. L, M and S refer to the long-, mid- and short-styled morphs, respectively. The arrows indicate cross-pollinations between anthers and stigmas of equivalent height that are typical in most tristylous populations. With this mating pattern a 1:1:1 ratio of floral morphs is expected at equilibrium as a result of frequency dependent mating. Tristyly is governed by two diallelic Mendelian loci (S, M), with the S-locus epistatic to the M-locus.
The first part of my research has been to assemble the genome of E. paniculata, as a genome assembly is required for gene mapping and other projects. I have obtained DNA sequences from E. paniculata using two cutting-edge sequencing techniques: PacBio and 10X Chromium. With these sequences, we have conducted de novo genome assemblies with two software programs, SMARTdenovo and Canu. I then merged the two assemblies with several software programs including quickmerge. After the assemblies were merged, I then obtained a chromosome-level (i.e. dozens of megabases long) assembly using Hi-C sequencing.
To find the genes underlying tristyly and how they are arranged in the genome, I have been attempting to identify genes that show an association with each of the floral morphs. I have recently obtained Illumina sequences of 60 plants (20 of each floral morph) and am conducting association mapping with the DNA sequences and several software programs including freebayes to identify SNPs that co-segregate with each of the floral morphs.
In parallel and to characterize the function of each gene, i.e. which gene controls which part of the tristylous syndrome, I have conducted floral organ-specific transcriptomic analysis. I have dissected hundreds of floral buds, collected styles and stamens of each morph at specific developmental stages, and extracted RNA from them. I am now analyzing sequencing reads from each RNA sample, assembling transcripts, and searching for genes only expressed in certain organ types of specific morphs. The results should help me identify genes controlling individual components of the tristylous syndrome.
After I identify the tristyly genes, I plan to conduct virus-induced gene silencing for each of the genes to verify their functions. I will also use molecular dating techniques to estimate the time when the S and M loci originated.
Publications
Jiao, M., Xue, H., Yan, J., Zheng, Z., Wang, J., Zhao, C., Zhang, L. & Zhou, W. (2021). Tree abundance, diversity and their driving and indicative factors in Beijing’s residential areas. Ecological Indicators, 125: 107462.
Xue H., Zhao T., Chen H. (2018). Translation of McGhee K. and Mckay G. Children’s Encyclopedia of Animals. Original work published 2014. Post Wave, Beijing, China.
Xue H.R.*, Yamaguchi N.,* Driscoll C.A.,*, Han Y.,*, Bar-Gal G.K., Zhuang Y., Mazak J.H., Macdonald D.W., O’Brien S.J., and Luo S.J. (2015). Genetic ancestry of the extinct Javan and Bali tigers. Journal of Heredity, 106: 247–257. (*equal contribution)
Xue H.*, Shi H.*, Yu, Z.*, He S.*, Liu S., Hou, Y., Pan X., Wang H., Zheng P., Cui C., Viets H., Liang J., Zhang Y., Chen S., Zhang H.M. and Ouyang Q. (2014). Design, construction, and characterization of a set of biosensors for aromatic compounds. ACS Synthetic Biology 3: 1011–1014. (*equal contribution)